Since the dawn of the global SARS-CoV-2 pandemic, the need for unique and novel methodologies enabling viral detection has skyrocketed. As a result, various aspects of the traditional detection workflow were reworked and re-implemented to drive down cost, increase efficiency and decrease turn-around time to results. We saw this model applied to various detection-based assays across the market, from the development of several at-home lateral flow-based rapid tests to the development of molecular-detection assays that eliminate viral RNA extraction steps. Deviating from the traditional nasopharyngeal (NP) or oropharyngeal (OP) swabs as a sample type, some tests even adopted saliva as a novel sample type in molecular detection assays.
Keeping with the idea of innovation, the OMNI R&D team evaluated the role that bead mill homogenization played in an RT-qPCR viral detection workflow during the initial sample preparation steps. Preliminary proof-of-concept experiments showed lysis efficiency of the Bead Ruptor EliteTM Bead Mill Homogenizer in lysing HCoV-229E off NP swabs, releasing analytes into solution for direct-to-PCR detection. That’s right, an extraction-free RT-qPCR workflow (1). Eventually, moving to a SARS-CoV-2 based experiment model, this workflow continued to show promise, especially in an environment where extraction reagents and lab plastics were hard to come by.
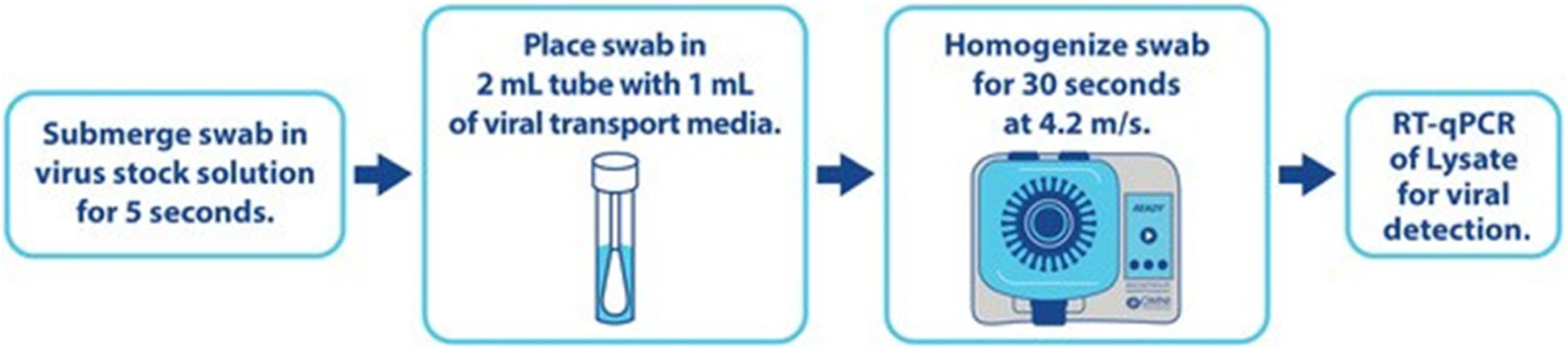
One collaborator from the University of Malawi College of Medicine deployed the direct-to-PCR SARS-CoV-2 workflow compared to standard extraction-based RT-qPCR detection from NP/OP swabs. In a resulting joint publication outlining the study, the vital importance of an extraction-free SARS-CoV-2 detection workflow in such resource-challenged areas is depicted by illuminating the following (2):
• A high agree ability in detection between direct-to-PCR and extraction-based methods.
• NP/OP sample processing time was reduced to 30 seconds using the Bead Ruptor EliteTM Bead Mill Homogenizer compared to 1 hour when using standard extraction-based methods.
• A reduction in materials and instruments needed to perform the assay.
Further down the line, saliva arose as an alternative sample type and potential workaround to sample collection hurdles posed by NP/OP swabs. Additionally, saliva was explored to potentially resolve the safety risk posed by healthcare workers in handling patient NP/OP swabs. As a result, many studies began popping up, concluding that an equivalent or higher level of sensitivity in SARS-CoV-2 detection from saliva was observed, supporting the rationale for change (3-7). With the change, however, comes new obstacles. Saliva is a viscous matrix that is not easily transferred via manual or automated pipetting techniques, which created an issue in the sample handling workflow. The Bead Ruptor EliteTM Homogenizer was recruited once again by several in-house researchers at OMNI and in the scientific community. Depicted in an end-to-end workflow, a 30-second homogenization step in our specialized 2 mL bead tube provided the following benefits:
• Reduced viscosity of saliva post-homogenization steps on the Bead Ruptor EliteTM Homogenizer.
• Hassle-free liquid handling into downstream extraction on the chemagic 360 insrument.
• Increased sensitivity of RT-qPCR assay amplifying SARS-CoV-2 genes of interest when compared to NP samples (8).
APPLICATION NOTE: SARS-CoV-2 Detection in Saliva Using Bead Beating Homogenization and a RT-PCR Test Kit.
As methods and sample types evolved, time went on, and reported cases of SARS-CoV-2 decreased (9). Next, the focus shifted to how we, as a scientific community, can apply what was applied to SARS-CoV-2 to other respiratory assays. Finally, attention shifted to the influenza A virus (IAV), which represents a significant disease burden in the United States each year and warrants innovation in detecting this endemic respiratory pathogen. To support needs centered around disease research, the OMNI R&D team set out to show the utility of the Bead Ruptor EliteTM Homogenizer again. In a comparison between standard extraction-based and direct-to-PCR, IAV was disrupted off NP swabs in a 30-second bead beating step, providing proof-of-concept for viral lysis upstream of RT-qPCR detection of matrix gene.
APPLICATION NOTE: Utilization of a Mechanical Homogenization-Based Direct-to-PCR Method for Influenza A Virus Detection
With intelligent and efficient sample preparation, downstream methods are enabled, empowering the relentless innovation of researchers, technicians, students, and other Bead Ruptor EliteTM Homogenizer users alike.
For more information on OMNI sample preparation technology, reach out to sales@omni-inc.com
For research use only. Not for use in diagnostic procedures.
Citations:
1. Morehouse, Z. P., Proctor, C. M., Ryan, G. L., & Nash, R. J. (2020). A novel two-step, direct-to-PCR method for virus detection off swabs using human coronavirus 229E. Virology journal, 17(1), 129. https://doi.org/10.1186/s12985-020-01405-y
2. Morehouse, Z. P., Samikwa, L., Proctor, C. M., Meleke, H., Kamdolozi, M., Ryan, G. L., Chaima, D., Ho, A., Nash, R. J., & Nyirenda, T. S. (2021). Validation of a direct-to-PCR COVID-19 detection protocol utilizing mechanical homogenization: A model for reducing resources needed for accurate testing. PloS one, 16(8), e0256316. https://doi.org/10.1371/journal.pone.0256316
3. To, K. K., Tsang, O. T., Leung, W. S., Tam, A. R., Wu, T. C., Lung, D. C., Yip, C. C., Cai, J. P., Chan, J. M., Chik, T. S., Lau, D. P., Choi, C. Y., Chen, L. L., Chan, W. M., Chan, K. H., Ip, J. D., Ng, A. C., Poon, R. W., Luo, C. T., Cheng, V. C., … Yuen, K. Y. (2020). Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. The Lancet. Infectious diseases, 20(5), 565–574. https://doi.org/10.1016/S1473-3099(20)30196-1
4. To, K. K., Tsang, O. T., Yip, C. C., Chan, K. H., Wu, T. C., Chan, J. M., Leung, W. S., Chik, T. S., Choi, C. Y., Kandamby, D. H., Lung, D. C., Tam, A. R., Poon, R. W., Fung, A. Y., Hung, I. F., Cheng, V. C., Chan, J. F., & Yuen, K. Y. (2020). Consistent Detection of 2019 Novel Coronavirus in Saliva. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America, 71(15), 841–843. https://doi.org/10.1093/cid/ciaa149
5. Azzi, L., Carcano, G., Gianfagna, F., Grossi, P., Gasperina, D. D., Genoni, A., Fasano, M., Sessa, F., Tettamanti, L., Carinci, F., Maurino, V., Rossi, A., Tagliabue, A., & Baj, A. (2020). Saliva is a reliable tool to detect SARS-CoV-2. The Journal of infection, 81(1), e45–e50. https://doi.org/10.1016/j.jinf.2020.04.005
6. Yoon, J. G., Yoon, J., Song, J. Y., Yoon, S. Y., Lim, C. S., Seong, H., Noh, J. Y., Cheong, H. J., & Kim, W. J. (2020). Clinical Significance of a High SARS-CoV-2 Viral Load in the Saliva. Journal of Korean medical science, 35(20), e195. https://doi.org/10.3346/jkms.2020.35.e195
7. Wyllie, A. L., Fournier, J., Casanovas-Massana, A., Campbell, M., Tokuyama, M., Vijayakumar, P., Warren, J. L., Geng, B., Muenker, M. C., Moore, A. J., Vogels, C. B. F., Petrone, M. E., Ott, I. M., Lu, P., Venkataraman, A., Lu-Culligan, A., Klein, J., Earnest, R., Simonov, M., Datta, R., … Ko, A. I. (2020). Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. The New England journal of medicine, 383(13), 1283–1286. https://doi.org/10.1056/NEJMc2016359
8. Sahajpal, N. S., Mondal, A. K., Ananth, S., Njau, A., Ahluwalia, P., Kota, V., Caspary, K., Ross, T. M., Farrell, M., Shannon, M. P., Fulzele, S., Chaubey, A., Hegde, M., Rojiani, A. M., & Kolhe, R. (2021). Clinical Validation of a Sensitive Test for Saliva Collected in Healthcare and Community Settings with Pooling Utility for Severe Acute Respiratory Syndrome Coronavirus 2 Mass Surveillance. The Journal of molecular diagnostics: JMD, 23(7), 788–795. https://doi.org/10.1016/j.jmoldx.2021.04.005
9. Centers for Disease Control. (n.d.). CDC Covid Data tracker. Centers for Disease Control and Prevention. Retrieved February 10, 2023, from https://covid.cdc.gov/covid-data-tracker/#datatracker-home
10. Centers for Disease Control and Prevention & National Center for Immunization and Respiratory Diseases. (2022, October 4). Disease burden of flu. Centers for Disease Control and Prevention. Retrieved February 10, 2023, from https://www.cdc.gov/flu/about/burden/index.html


